Part:BBa_K3846304
OleT-Fd expression cassette (rhamnose inducible) - cyanobacteria
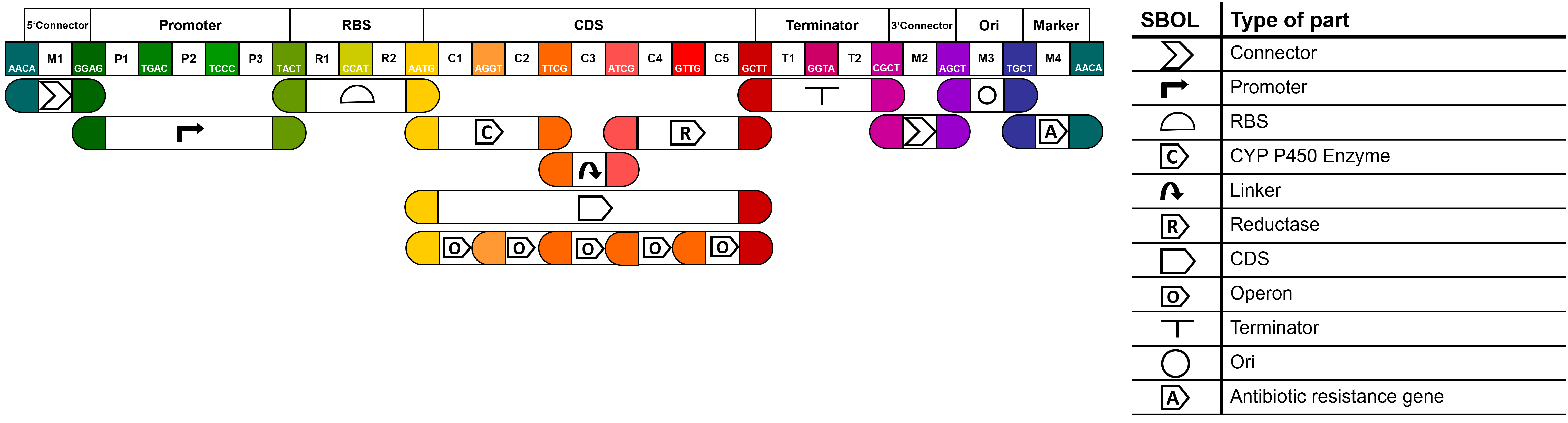
Our project focuses on enzymatic reactions catalysed by cytochrome P450 enzymes. This group of enzymes always rely on a reductase to be functional. To increase the activity of the P450 enzyme we decided to create fusion proteins, consisting of an N-terminal cytochrome P450 enzyme and a C-terminal NADPH-cytochrome P450 reductase connected by a linker sequence. It is likely that the composition of the linker sequence influences the activity of the fusion protein since electrons have to be transferred from the reductase to the P450 enzyme and the structural arrangement of the domains should have an effect on this process. Optimising the linker sequence is therefore crucial for reaching optimal enzyme activity. To allow modular combinations of different P450 enzymes, reductases and linkers we designed these parts interchangeably using the following MoClo compatible fusion sites.
fusion sites:
- CYP P450 enzyme (AATG -TTCG)
- linker sequence (TTCG - ATCG)
- reductase (ATCG - GCTT)
In this inducible expression cassette for Synechocystis PCC 6803 a CYP P450-reductase fusion protein is expressed under the rhamnose-inducible rhaA promoter. The transcription activator rhaS is also present. Recent studies have shown that the rhamnose-inducible can achieve an up to 55x fold induction and is very suitable to be used for the expression of metabolic high burden fusion proteins.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 15
Illegal NheI site found at 38
Illegal NheI site found at 1070
Illegal NheI site found at 1093 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 2452
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
iGEM Hamburg 2021 part collection
Terpenoids are an important group of natural products used as biofuels, drugs or fragrances. Naturally occuring in plants it has been shown that microbial terpene production in microorganisms like yeast, E. coli or cyanobacteria is possible. Nevertheless iGEM projects seem to rarely focus on this interesting class of natural products which is correlated with a lack of useful parts inside the iGEM registry.
Fortunately we were able to change that and designed a novel golden gate based toolbox which allows.
- production of terpenoid precursors and simple terpenoids
- creation of CYP P450-reductase fusion enzymes to optimise processing of terpenoid precursors and production of bioactive target products
- modularity of the system to enable exchange of linker sequences/promoters/etc. (MoClo-compatible toolbox)
MoClo-based Part Design 2.0
To improve the usefulness of our parts, we then aimed to make our parts compatible with the MoClo standard of goten gate based IIS restriction enzyme assembly. Thereby we expanded the Common Genetic Syntax for fusion sites to allow the creation of a) fusion proteins connected by linker sequences and b) multiple CDS expressed in an operon. More useful information and an overview of all our parts can be found on our wiki.
| None |